Hot Topics #25 (June 3, 2024)
Transformers can do arithmetic, designing and scaffolding proteins, diffusion LMs, and more.

Transformers Can Do Arithmetic with the Right Embeddings: McLeish et al.: May 27, 2024
Abstract: The poor performance of transformers on arithmetic tasks seems to stem in large part from their inability to keep track of the exact position of each digit inside of a large span of digits. We mend this problem by adding an embedding to each digit that encodes its position relative to the start of the number. In addition to the boost these embeddings provide on their own, we show that this fix enables architectural modifications such as input injection and recurrent layers to improve performance even further.
With positions resolved, we can study the logical extrapolation ability of transformers. Can they solve arithmetic problems that are larger and more complex than those in their training data? We find that training on only 20 digit numbers with a single GPU for one day, we can reach state-of-the-art performance, achieving up to 99% accuracy on 100 digit addition problems. Finally, we show that these gains in numeracy also unlock improvements on other multi-step reasoning tasks including sorting and multiplication.
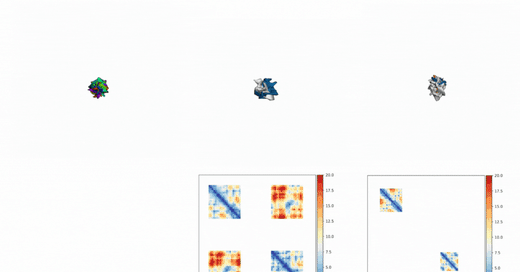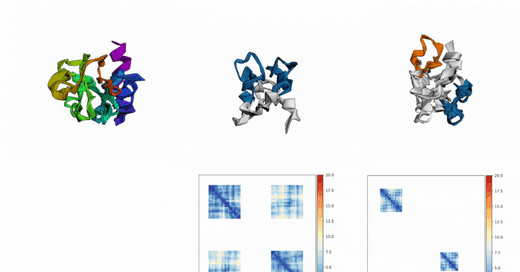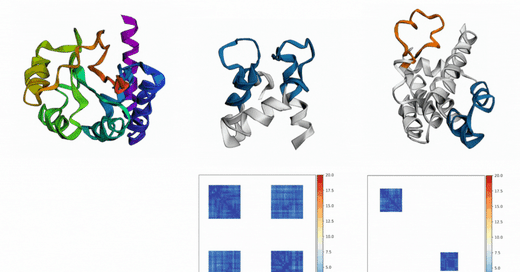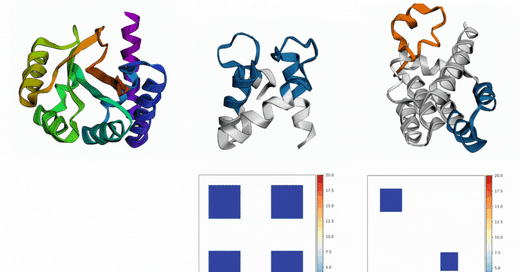
Out of Many, One: Designing and Scaffolding Proteins at the Scale of the Structural Universe with Genie 2: Lin et al.: May 24, 2024
Abstract: Protein diffusion models have emerged as a promising approach for protein design. One such pioneering model is Genie, a method that asymmetrically represents protein structures during the forward and backward processes, using simple Gaussian noising for the former and expressive SE(3)-equivariant attention for the latter. In this work we introduce Genie 2, extending Genie to capture a larger and more diverse protein structure space through architectural innovations and massive data augmentation. Genie 2 adds motif scaffolding capabilities via a novel multi-motif framework that designs co-occurring motifs with unspecified inter-motif positions and orientations. This makes possible complex protein designs that engage multiple interaction partners and perform multiple functions. On both unconditional and conditional generation, Genie 2 achieves state-of-the-art performance, outperforming all known methods on key design metrics including designability, diversity, and novelty. Genie 2 also solves more motif scaffolding problems than other methods and does so with more unique and varied solutions. Taken together, these advances set a new standard for structure-based protein design. Genie 2 inference and training code, as well as model weights, are freely available at: this https URL.
Diffusion Language Models Are Versatile Protein Learners: Wang et al.: February 28, 2024
Abstract: This paper introduces diffusion protein language model (DPLM), a versatile protein language model that demonstrates strong generative and predictive capabilities for protein sequences. We first pre-train scalable DPLMs from evolutionary-scale protein sequences within a generative self-supervised discrete diffusion probabilistic framework, which generalizes language modeling for proteins in a principled way. After pre-training, DPLM exhibits the ability to generate structurally plausible, novel, and diverse protein sequences for unconditional generation. We further demonstrate the proposed diffusion generative pre-training makes DPLM possess a better understanding of proteins, making it a superior representation learner, which can be fine-tuned for various predictive tasks, comparing favorably to ESM2 (Lin et al., 2022). Moreover, DPLM can be tailored for various needs, which showcases its prowess of conditional generation in several ways: (1) conditioning on partial peptide sequences, e.g., generating scaffolds for functional motifs with high success rate; (2) incorporating other modalities as conditioner, e.g., structure-conditioned generation for inverse folding; and (3) steering sequence generation towards desired properties, e.g., satisfying specified secondary structures, through a plug-and-play classifier guidance.
RNAFlow: RNA Structure & Sequence Design via Inverse Folding-Based Flow Matching: Nori & Jin: May 29, 2024
Abstract: The growing significance of RNA engineering in diverse biological applications has spurred interest in developing AI methods for structure-based RNA design. While diffusion models have excelled in protein design, adapting them for RNA presents new challenges due to RNA's conformational flexibility and the computational cost of fine-tuning large structure prediction models. To this end, we propose RNAFlow, a flow matching model for protein-conditioned RNA sequence-structure design. Its denoising network integrates an RNA inverse folding model and a pre-trained RosettaFold2NA network for generation of RNA sequences and structures. The integration of inverse folding in the structure denoising process allows us to simplify training by fixing the structure prediction network. We further enhance the inverse folding model by conditioning it on inferred conformational ensembles to model dynamic RNA conformations. Evaluation on protein-conditioned RNA structure and sequence generation tasks demonstrates RNAFlow's advantage over existing RNA design methods.
Geometry-Informed Neural Networks: Berzins et al.: May 27, 2024
Abstract: Geometry is a ubiquitous language of computer graphics, design, and engineering. However, the lack of large shape datasets limits the application of state-of-the-art supervised learning methods and motivates the exploration of alternative learning strategies. To this end, we introduce geometry-informed neural networks (GINNs) to train shape generative models \emph{without any data}. GINNs combine (i) learning under constraints, (ii) neural fields as a suitable representation, and (iii) generating diverse solutions to under-determined problems. We apply GINNs to several two and three-dimensional problems of increasing levels of complexity. Our results demonstrate the feasibility of training shape generative models in a data-free setting. This new paradigm opens several exciting research directions, expanding the application of generative models into domains where data is sparse.
A Recipe for Charge Density Prediction: Fu et al.: May 29, 2024
Abstract: In density functional theory, charge density is the core attribute of atomic systems from which all chemical properties can be derived. Machine learning methods are promising in significantly accelerating charge density prediction, yet existing approaches either lack accuracy or scalability. We propose a recipe that can achieve both. In particular, we identify three key ingredients: (1) representing the charge density with atomic and virtual orbitals (spherical fields centered at atom/virtual coordinates); (2) using expressive and learnable orbital basis sets (basis function for the spherical fields); and (3) using high-capacity equivariant neural network architecture. Our method achieves state-of-the-art accuracy while being more than an order of magnitude faster than existing methods. Furthermore, our method enables flexible efficiency-accuracy trade-offs by adjusting the model/basis sizes.
SaprotHub: Making Protein Modeling Accessible to All Biologists: This is the Colab version of SaProt, a pre-trained protein language model designed for various downstream protein tasks.
ColabSaprot is a platform where Protein Language Models(PLMs) are more accessible and user-friendly for biologists, enabling effortless model training and sharing within the scientific community.
We've established the SaprotHub for storing and sharing models and datasets, where you can explore extensive collections for specific protein prediction tasks.
We hope ColabSaprot and SaprotHub can contribute to advancing biological research, fostering collaboration, and accelerating discoveries in the field. You can access our paper for further details.
For detailed steps of each section, please refer to the manual.
Genie 2 code: This repository provides the implementation code for our preprint, including training and inference code, as well as model weights. For the in-silico evaluation pipeline, which is used to assess the designability, diversity and novelty of our generated structures, we provide them in a seperate repository since it is independent of Genie 2 and could be applicable for evaluating other protein structure diffusion models.